IMET scientists crack blue crab’s genetic code
Depending on the day, you may find Dr. J. Sook Chung caring for baby blue crabs in IMET’s Aquaculture Research Center, advising a student or postdoc, or teaching kids to make a headband with crab eyes from construction paper. Whatever she is doing, she is always enthusiastic about it. So when Dr. Chung, who goes by Sook, had the idea to sequence the blue crab’s genome, she dove in headfirst and her passion proved infectious. She knew this information would prove indispensable to science research and to the management of the blue crab fishery, which is economically and culturally significant to Maryland.
Dr. Tsvetan Bachvaroff, who goes by Tsetso, was the first to join Sook’s mission. “About five years ago, Sook came to my office and asked whether I would be willing to and capable of sequencing the blue crab genome. We both realized what a major commitment this would be,” he said. “And we thought that we could meet that challenge.” The task certainly was daunting, with an estimated genome size of one billion bases and just two researchers devoted to the project.

It was important for Sook to bring Tsetso onto the team because of his deep knowledge of bioinformatics, the science of complex biological information like genomes. Tsetso has taught this subject to graduate students for several years and has worked with the genetic information of creatures like dinoflagellates and marine sponges. Still, the blue crab genome was much bigger than anything else he had sequenced before. To round out the group of scientists, Dr. Louis Plough joined with his expertise in population genetics.
With Louis and Tsetso on board, Sook knew she had a group capable of sequencing the blue crab genome, but she still had to convince someone to fund the project. The Blue Crab Genome Initiative, as they named it, is unique in that it is funded primarily by private donors. These private citizens became advocates of the project, demonstrating its significance to the public. According to our lead private donors, Mike and Trish Davis, "Sook’s enthusiasm for this work was contagious, and we were thrilled to discover that the skills and technology for this exciting project existed right here in Baltimore. Her analogy of requiring a 'blueprint' for the blue crab, to really understand it, just like builders use blueprints to understand a building clearly resonated with the donors, and we all really wanted this scientific milestone achieved here in Maryland."
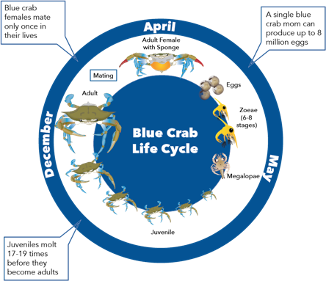
With a great team of scientists and the generous support of donors, the Blue Crab Genome Initiative needed one more team member – a blue crab! In late October 2018, Sook went out on the Chesapeake Bay on a crabber’s boat and collected dozens of young female blue crabs, which she kept as broodstock (animals that produce offspring) in IMET’s Aquaculture Research Center (ARC). This was possible because of research done at IMET several years ago as part of the
Blue Crab Advanced Research Consortium to close the blue crab lifecycle in captivity – the first time this was done in North America. One of the females she brought back became an adult, mated, and produced offspring, all within IMET’s ARC. One of these ARC-born offspring also produced juveniles. This crab was perfect – she had roots in the Chesapeake, was raised at IMET, and had demonstrated that she had good genes for reproducing. That’s why Sook and Tsetso refer to her as “the Chosen One.”
Sook and Tsetso also referred to this crab as the Chosen One to avoid confusion. Sook is the name for a female crab, so Blue Crab Genome Initiative now had two Sooks. When asked about this strange coincidence, Sook (the scientist) responded, “I may be born, in South Korea, to study blue crabs in Maryland.”
Sook began the work that she was destined to do, isolating DNA from the Chosen One and sending it off to be sequenced. Sequencing will return reads, or fragments of A, C, G, and T, the four bases that form DNA and that therefore create the basis of all life. Sook and Tsetso used multiple methods of DNA sequencing, ultimately generating data on 254 billion bases. The initial step of assembly, or establishing the correct order of every A, C, G, and T for the genome, required a special computer running night and day for over six months. Eventually, the scientists determined that blue crabs have 40-50 chromosomes, double the amount in humans. These chromosomes are relatively short, resulting in a genome with about a billion bases, compared to humans’ 3 billion. Still, they have high gene density, with even more genes than humans.
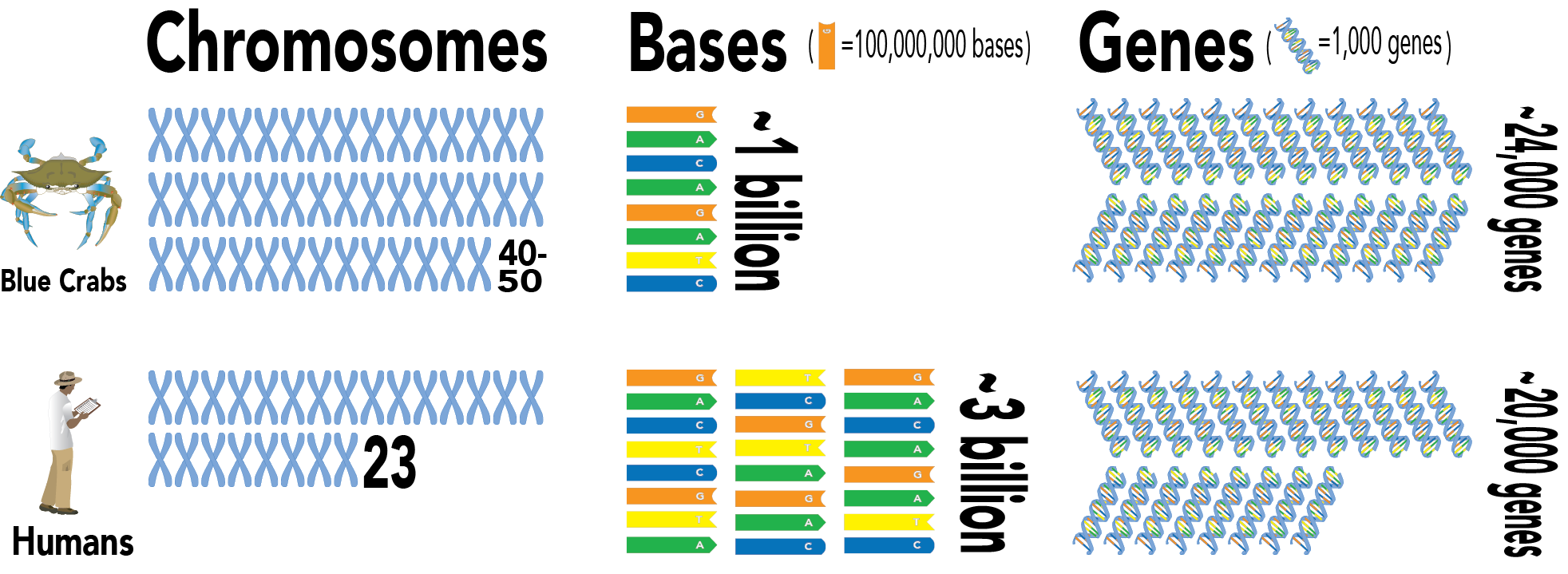
It is important to establish the correct order of bases because that sequence ultimately determines how an organism will grow and develop. Describing the challenge of this process, Tsetso said, “Imagine you take several volumes of an encyclopedia and you have a hundred copies of each volume. You put them all through a paper shredder and then you have to use that to reconstruct the original volumes of the encyclopedia.” Once the encyclopedia, or genome, is back in the correct order, you can begin to identify genes and use it like a reference book, looking up genes to answer questions.

The question that most interests Sook now is if she can identify genetic characteristics that enhance female blue crabs’ ability to reproduce. She was the lead scientist on closing the blue crab life cycle in the Aquaculture Research Center and so she observed generations of crabs mating, producing eggs, and releasing juveniles. “When you do that,” she said, “you find there’s huge variation in reproductive success depending on the female. So, as a scientist, I wanted to know, what’s the genetic make-up? What makes them successful or what makes them a failure in juvenile production?” The genome is a crucial tool in answering those questions.
The work Sook proposes is made possible by the availability of a complete genome, and it has many important applications. If we could characterize not just how many crabs are in the Bay, but also how likely they are to reproduce successfully, that could aid in fisheries policies, helping to maintain a healthy ecosystem and economy. If we could breed females who are particularly reproductively successful, that could help enable the production of blue crabs in aquaculture. Both of those outcomes lead to something beloved by Marylanders: blue crab on your plate, smothered in Old Bay.
The sequencing of the Blue Crab Genome was made possible by the support of the following generous donors: The G. Unger Vetlesen Foundation, Mike and Trish Davis , Don and Cathy MacMurray , James J. Albrecht , Bertram and Debbie Winchester, Arnold and Alison Richman, Maryland Sea Grant, Arthur Jib Edwards , Jum Sook Chung, Richard L. Franyo, Edward St. John Foundation , Tom and Nancy Reynolds , James E. Connell , Russell T. Hill , Bill and Chris Hufnell , David Balcom , J. Mitchell Neitzey , James and Jenny Corckran, Richard and Maureen Roden , Nicholas L. Hammond